Research
The majority of projects in the laboratory use crystallography to address fundamental issues in biomedical science: How is protein function defined by structure? How do protein cofactors modulate enzymes? How does structure prescribe the binding affinity of a metal?
To begin to dissect the intricate relationship between a protein cofactor and its enzyme we selected to study the prothrombinase complex in collaboration with Dr. Kenneth Mann (Biochemistry). The prothrombinase complex is composed the coagulation co-factor factor Va, a proteolytically activated form of factor V (330 kDa) and the active enzyme, factor Xa. The prothrombinase complex is responsible for the tremendous burst of thrombin necessary for the generation of a successful clot. As a start to providing a real structural understanding of cofactors we solved the structure of bovine factor Vai (Figure 1). While efforts continue to crystallize the complex (factor Va + factor Xa +/- prothrombin) we have more recently focused on computer modeling (Figure 2) as a method to generate hypotheses that can be tested in the laboratory.
In a tremendously successful collaboration with Dr. Anne B. Mason (Biochemistry) we have engaged in structural studies of transferrin, the major plasma protein responsible for iron transport, and its specific receptor, the transferrin receptor. Transferrin (80 kDa) is a glycoprotein with two homologous lobes. Each lobe consists of two domains that form a deep cleft which bind a single Fe (III) ion in conjunction with the concomitant binding of a carbonate ion in a pH dependent manner. We have been solving structures of single lobes (Figure 3), the entire transferrin molecule, as well as the transferrin/transferrin receptor complex (Figure 4) to understand the complex relationship between structure, pH and iron status (bound/not bound).
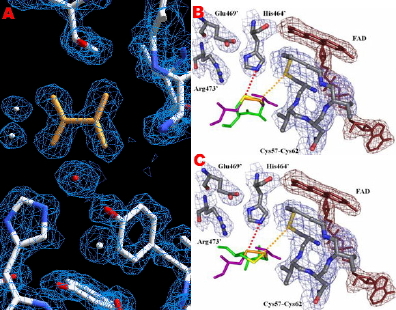
In collaboration with Dr. Robert Hondal (Biochemistry) we have been using structure to explore mechanism in thioredoxin reductases (TR). Our structure of Drosophila melanogaster TR allowed us to postulate why mammalian TRs requires selenocysteine but Drosophila TR can utilize cysteine (Figure 5).
In addition to the on-going structural work in the laboratory, we maintain an active collaboration in the mathematical modeling of coagulation with Kathleen Brummel-Ziedins (Biochemistry), Thomas Orfeo (Biochemistry), and Kenneth Mann (Biochemistry). Recent efforts have been directed at extending our “base” model (Figure 6).
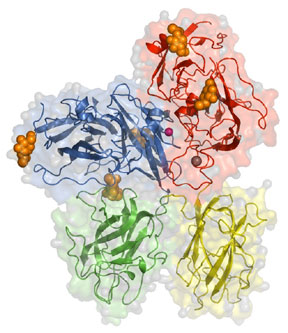
Crystal structure of bovine factor Vai (missing the A2 domain). Domains present are indicated, carbohydrates shown in orange, the copper atom is pink, and the calcium atom is in grey. For more information please see Adams et al. PNAS. 101: 8918-8923, 2004.
Molecular model of the human prothrombinase complex (factor Va and factor Xa) on a hypothetical membrane. Factor Va is colored as shown in Figure 1 with the exception that the A2 domain is colored orange. Factor Xa is shown in cyan with its active site indicated. For more information please see Everse et al. “A molecular model of the human prothrombinase complex” (2008) Recent Advances in Thrombosis and Hemostasis 2008. K. Tanaka & E.W. Davie (Eds). Springer Japan: Chiyoda-ku, pp. 107-132.