Genetic and Epigenetic Regulation of Gene Expression
 While genetic information encoded by DNA provides a framework for gene expression, epigenetic regulation is equally critical for physiological responsiveness to extracellular cues. Our laboratory pioneered examining the chromatin structure of several key gene loci (e.g., histone H4, osteocalcin, Runx2) to understand how architectural and topological constraints on gene promoters and enhancers at distal regulatory sites in chromatin contribute to epigenetic control of gene expression. These studies have evolved into detailed molecular analyses of histone modifications (e.g., H4 acetylation, H3 methylation), SWI/SNF related chromatin remodeling and genome-wide transcription factor binding analyses using ChIP-on-chip and ChIP-Seq approaches (see Nuclear Structure and Function).
While genetic information encoded by DNA provides a framework for gene expression, epigenetic regulation is equally critical for physiological responsiveness to extracellular cues. Our laboratory pioneered examining the chromatin structure of several key gene loci (e.g., histone H4, osteocalcin, Runx2) to understand how architectural and topological constraints on gene promoters and enhancers at distal regulatory sites in chromatin contribute to epigenetic control of gene expression. These studies have evolved into detailed molecular analyses of histone modifications (e.g., H4 acetylation, H3 methylation), SWI/SNF related chromatin remodeling and genome-wide transcription factor binding analyses using ChIP-on-chip and ChIP-Seq approaches (see Nuclear Structure and Function).
Beyond the conventional epigenetic mechanisms that include post-transcriptional modifications of histones, and methylation of CpG moieties in gene promoters, we have identified a unique epigenetic mechanism, where phenotypic transcription factors remain associated with target genes during mitosis. This association, termed architectural epigenetics (‘Bookmarking’) ensures that the progeny cells maintain the cell growth and proliferative potential, as well as remain committed to lineage of the parent cells. We are actively exploring mechanisms that ensure sustained architectural epigenetics during symmetrical division of committed cells and during asymmetrical mitosis of stem cells.
Another epigenetic mechanism that has become a focus of our research group is the involvement of non-coding RNAs, microRNAs, and long non-coding RNAs, in regulating osteoblast- and hematopoiesis-related gene expression. We have carried out genome-wide screens for the expression of miRs and long non-coding RNAs under various physiological conditions and have identified miR signatures that have the potential to predict a biological response or a pathological outcome. Our group is further dissecting roles of these non-coding RNA signatures in osteoblastogenesis (see Musculoskeletal Biology and Pathology) and hematopoeisis, as well as in human leukemia. We have identified a mitotically associated long non-coding RNA in triple-negative breast cancer cells designated MANCR that is required to stabilize the breast cancer-compromised genome. Based on our discovery that knock-out of MANCR in triple-negative breast cancer cells results in apoptosis/cell death we are investigating MANCR knock-out as a targeted strategy for triple-negative breast cancer in vivo utilizing a xenograft mouse mode.
Our research group was among the first to investigate cancer-compromised epigenetic control of gene expression during the onset of progression of breast cancer. In a tumor progression breast cancer cell culture model we have leverage chromosome conformation capture approaches to identify cancer-compromised modifications in higher-order chromatin organization. We have directly confirmed breast cancer-compromised inter and intrachromosomal interaction by multispectral imaging. We are mechanistically defining epigenetic control of aberrant chromatin organization in breast cancer cells by genomic strategies that identify histone modifications and DNA methylation. Initial epigenetic responses to degron ablation of the RUNX1 tumor suppressor in mammary epithelial cells is providing the first direct indication of regulatory consequences that are functionally linked with breast cancer initiation. We are pursuing clinically relevant validation of our findings by multi-omic and spatial transcriptomic analysis of patient-derived breast tumors.
Landmark Papers
Tye CE, Ghule PN, Gordon JAR, Kabala FS, Page NA, Falcone MM, Tracy KM, van Wijnen AJ, Stein JL, Lian JB, Stein GS. LncMIR181A1HG is a novel chromatin-bound epigenetic suppressor of early stage osteogenic lineage commitment. Sci Rep. 2022 May 11;12(1):7770. doi: 10.1038/s41598-022-11814-4. PMID: 35546168; PMCID: PMC9095685.
Gordon JAR, Tye CE, Banerjee B, Ghule PN, van Wijnen AJ, Kabala FS, Page NA, Falcone MM, Stein JL, Stein GS, Lian JB. LINC01638 sustains human mesenchymal stem cell self-renewal and competency for osteogenic cell fate. Sci Rep. 2023 Nov 20;13(1):20314. doi: 10.1038/s41598-023-46202-z. PMID: 37985890; PMCID: PMC10662126.
Ghule PN, Boyd JR, Kabala F, Fritz AJ, Bouffard NA, Gao C, Bright K, Macfarlane J, Seward DJ, Pegoraro G, Misteli T, Lian JB, Frietze S, Stein JL, van Wijnen AJ, Stein GS. Spatiotemporal higher-order chromatin landscape of human histone gene clusters at histone locus bodies during the cell cycle in breast cancer progression. Gene. 2023 Jul 1;872:147441. doi: 10.1016/j.gene.2023.147441. Epub 2023 Apr 23. Erratum in: Gene. 2023 May 11;873:147469. PMID: 37094694; PMCID:
Fritz AJ, Ghule PN, Toor R, Dillac L, Perelman J, Boyd J, Lian JB, Gordon JAR, Frietze S, Van Wijnen A, Stein JL, Stein GS. Spatiotemporal Epigenetic Control of the Histone Gene Chromatin Landscape during the Cell Cycle. Crit Rev Eukaryot Gene Expr. 2023;33(3):85-97. doi: 10.1615/CritRevEukaryotGeneExpr.2022046190. PMID: 37017672; PMCID: PMC10826887.
Messier TL, Boyd JR, Gordon JAR, Tye CE, Page NA, Toor RH, Zaidi SK, Komm BS, Frietze S, Stein JL, Lian JB, Stein GS. Epigenetic and transcriptome responsiveness to ER modulation by tissue selective estrogen complexes in breast epithelial and breast cancer cells. PLoS One. 2022 Jul 21;17(7):e0271725. doi: 10.1371/journal.pone.0271725. PMID: 35862394; PMCID: PMC9302754. PMC10370284.
Hong D, Fritz AJ, Zaidi SK, van Wijnen AJ, Nickerson JA, Imbalzano AN, Lian JB, Stein JL, Stein GS. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol. 2018 Dec;233(12):9136-9144. doi: 10.1002/jcp.26847. Epub 2018 Jul 3. PMID: 29968906; PMCID: PMC6185773.
Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control.Nat Rev Genet. 2010 Aug;11(8):583-9.
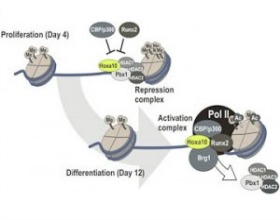
Gordon JA, Hassan MQ, Saini S, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol. 2010 Jul;30(14):3531-41.
Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008 May 6;105(18):6632-7.
Gutierrez J, Paredes R, Cruzat F, Hill DA, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling by SWI/SNF results in nucleosome mobilization to preferential positions in the rat osteocalcin gene promoter. J Biol Chem. 2007 Mar 30;282(13):9445-57.
Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3189-94. Epub 2007 Feb 20.
Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007 Jan 25;445(7126):442-6.
Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):14852-7.
Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999 Nov;19(11):7491-500.
Montecino M, Frenkel B, van Wijnen AJ, Lian JB, Stein GS, Stein JL. Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry. 1999 Jan 26;38(4):1338-45.